Degradation of PET Nanoplastic Oligomers at the Novel PHL7 Target:Insights from Molecular Docking and Machine Learning
Keywords:
Polyethylene terephthalate, Nanoplastic, Polyester Hydrolase Leipzig 7, binding affinity, Artificial Neutral NetworkAbstract
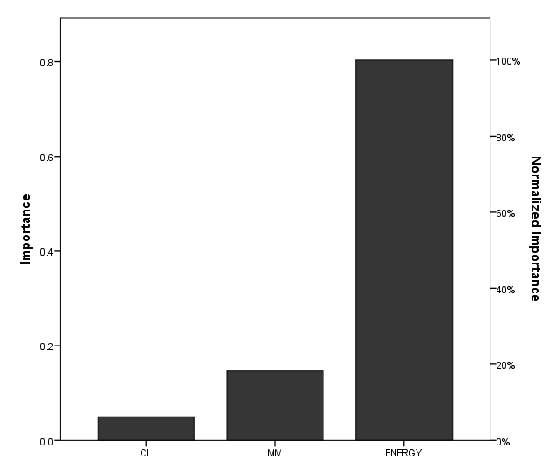
The versatility of Polyethylene terephthalate (PET) as a material with numerous applications in the food industry and its recalcitrance to chemical and microbial degradation has recently made it an environmental nuisance. In this study, we applied computational methods to ascertain the dependence of PET nanoplastic (NP) degradation on the chain length of the oligomer. The binding affinities of the NPs on the novel enzyme Polyester Hydrolase Leipzig 7 (PHL7) were used to relate their ease of degradation at the enzyme active site. The results revealed that the binding affinity of PET NPs at the enzyme target decreased from -5.2 kcal/mol to -0.8 kcal/mol, with an increase in PET chain length from 2.18 nm to 5.45 nm (2-5 PET chains). The binding affinities became positive at chain lengths 6.54 nm (6 PET chains) and above. These findings indicated that PET NP degradation at this enzyme’s active site is most efficient as chain length decreases from 5-2 units and is not likely to occur at longer PET chains. A feedforward Artificial Neutral Network (ANN) analysis predicted that the energy of the PET NPs is a very important factor in its degradation.
Published
How to Cite
Issue
Section
Copyright (c) 2023 C. E. Duru, C. E. Enyoh, I. A. Duru, M. C. Enedoh

This work is licensed under a Creative Commons Attribution 4.0 International License.




